Pre-processing of TLS or MLS point clouds for deriving size-distance-dependent competition indizes
For extracting information on tree level from close-range laser scanning (TLS or MLS), you can use e.g. the TreeLS package. We will show a workflow on how to implement the output of TreeLS to derive inventory based competition indices within TreeCompR.
First, load the .las data, normalize the height, sample the point cloud, estimate tree occurrence regions, classify stem points and estimate the diameter and height of each individual tree.
file <- "path/to/file"
tls <- readTLS(file)
tls_norm <- tlsNormalize(tls, keep_ground = F)
thin <- tlsSample(tls_norm, smp.voxelize(0.01))
map <- treeMap(thin, map.hough(min_density = 0.1), 0)
tls_t <- treePoints(tls_norm, map, trp.crop())
tls_stem <- stemPoints(tls_t, stm.hough())
inv <- tlsInventory(tls_stem, d_method=shapeFit(shape='circle',
algorithm = 'ransac'))
inv$dbh <- (inv$Radius * 2) %>% select(TreeID, X, Y, H, dbh)
head(inv)
#> TreeID X Y H dbh
#> 1: 1 11.709633 3.0399174 24.72817 1.06845322
#> 2: 2 11.953612 2.3900734 24.98053 0.32118377
#> 3: 3 7.429792 -4.9559437 23.75091 0.18467989
#> 4: 4 7.150391 5.7965706 24.05617 0.94364591
#> 5: 5 7.088778 0.8337127 24.07538 0.16088486
#> 6: 6 7.184398 5.2539175 23.94043 0.07801985 Use extracted tree information within TreeCompR
To ensure that the inventory data is assigned correctly, check the
data and make sure that you specify the units dbh and height if they
differ from the default, which is cm for dbh and m for height. Use
read_inv() to validate your inventory data within
TreeCompR. To visually check for which trees the CIs will
be quantified depending on the chosen search radius, you can use
define_target() and plot_target(). We
recommend to use target_source = "buff_edge" to
automatically calculate the CI for all trees in the plot except for
those that are less than one search radius away from the forest edge.
This is specifically important for TLS/MLS data, since these data types
usually only cover small forest plots rather than whole forests. In our
example, with a search radius of 10 m, only one tree is assigned as a
target tree.
#validate the inventory dataset "inv"
inventory <- read_inv(inv_source = inv, dbh_unit = "m", height_unit = "m")
#define the target tree(s) depending on the search radius
targets <- define_target(inv = inventory, target_source = "buff_edge",
radius = 10)
#>---------------------------------------------------------------
#> 'target_inv' class inventory dataset with defined target trees:
#> collection of 31 observations
#> Source of target trees: buffer around edge
#> ---------------------------------------------------------------
#> id x y dbh height target
#> 1 1 12.039 3.393 0.89 24.728 FALSE
#> 2 2 11.931 2.376 0.261 24.981 FALSE
#> 3 3 7.424 -4.956 0.19 23.751 FALSE
#> ... ... ... ... ... ...
#> 29 35 -6.407 -4.719 0.06 19.791 FALSE
#> 30 36 -7.484 -7.635 0.216 22.478 FALSE
#> 31 38 -5.287 1.051 0.06 22.923 FALSE
#>
# plot the target tree(s)
plot_target(targets)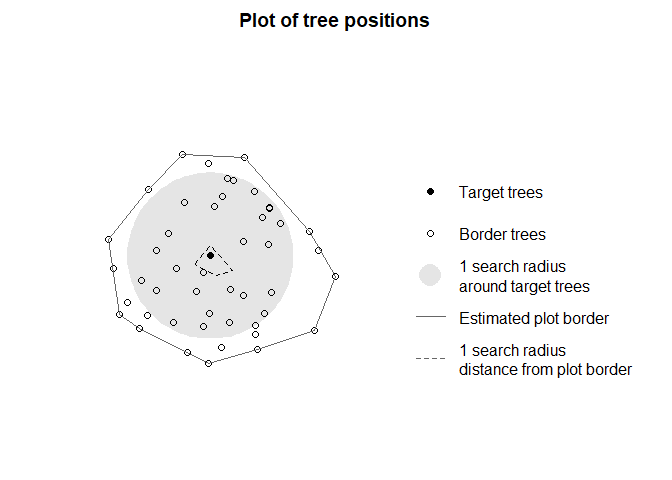
#example for calculating all possible CIs based on the parameters height and dbh
#with a search radius of 7m
compete_inv(inventory, target_source = "buff_edge", radius = 7,
method = "all_methods")
#> The following columns were used to create the inventory dataset:
#> id --- TreeID
#> x --- X
#> y --- Y
#> dbh --- dbh
#> height --- H
#>
#> ---------------------------------------------------------------------
#> 'compete_inv' class inventory with distance-based competition indices
#> Collection of data for 1 target and 30 edge trees.
#> Source of target trees: buffer around edge Search radius: 7
#> ---------------------------------------------------------------------
#> id ... dbh height CI_Hegyi CI_Braathe CI_RK1 CI_RK2 CI_RK3 CI_RK4
#> 18 20 ... 0.25 23.9 1.96 2.07 0.49 0.499 4.1 13.2Use cases
## If you focus on a specific CI, e.g. the widely used Hegyi-Index,
## you can choose it within methods = "CI_Hegyi".
compete_inv(inventory, target_source = "buff_edge", radius = 7,
method = "CI_Hegyi")
#> ------------------------------------------------------------
#> 'compete_inv' class distance-based competition index dataset
#> No. of target trees: 1 Source inventory size: 24 trees
#> Target source: 'buff_edge' (7 m) Search radius: 7 m
#> ------------------------------------------------------------
#> id x y dbh height CI_Hegyi
#> 1: 20 0.105 -0.492 24.961 23.871 1.986
## If you have a tree height column that is not automatically recognized within
## our validation of input data, you can set it manually:
## in read_inv():
inventory <- read_inv(inv_source = inventory, height = "h_column_name")
#> The following columns were used to create the inventory dataset:
#> id --- TreeID
#> x --- X
#> y --- Y
#> dbh --- dbh
#> height --- h_column_name
## or directly in compete_inv():
compete_inv(inventory2, target_source = "buff_edge", radius = 7,
height = "h_column_name", method = "CI_Hegyi")
#> The following columns were used to create the inventory dataset:
#> id --- TreeID
#> x --- X
#> y --- Y
#> dbh --- dbh
#> height --- h_column_name
#>
#> 7 trees outside the competitive zone around the target trees were removed.
#> 24 trees remain.
#> ------------------------------------------------------------
#> 'compete_inv' class distance-based competition index dataset
#> No. of target trees: 1 Source inventory size: 24 trees
#> Target source: 'buff_edge' (7 m) Search radius: 7 m
#> ------------------------------------------------------------
#> id x y dbh height CI_Hegyi
#> 1: 20 0.105 -0.492 0.25 23.871 1.986
## If your dbh is specified in cm, just change the dbh_unit to "cm"
compete_inv(inventory3, target_source = "buff_edge", radius = 7,
dbh_unit = "cm", method = "CI_Hegyi")
#> 7 trees outside the competitive zone around the target trees were removed.
#> 24 trees remain.
#> ------------------------------------------------------------
#> 'compete_inv' class distance-based competition index dataset
#> No. of target trees: 1 Source inventory size: 24 trees
#> Target source: 'buff_edge' (7 m) Search radius: 7 m
#> ------------------------------------------------------------
#> id x y dbh height CI_Hegyi
#> 1: 20 0.105 -0.492 24.961 23.871 1.986
## If you want to keep a column with certain user defined IDs for the trees,
## and it is not recognized, specify it with id = "name_column"
inventory <- read_inv(inv_source = inv4, id = "Tree_ID_User")
#> The following columns were used to create the inventory dataset:
#> id --- Tree_ID_User
#> x --- X
#> y --- Y
#> dbh --- dbh
#> height --- HIf you use other options to derive the tree parameters from TLS or
MLS data, you can customize read_inv() and
compete_inv() accordingly. For more examples, have a look
at some more examples.
