
Quantifying competition from point cloud data
Source:vignettes/competition-pointcloud.Rmd
competition-pointcloud.RmdGetting started
Before you can start computing competition indices based on point
cloud data from terrestrial or mobile laserscanning with
TreeCompR, these data have to be thoroughly vetted and
pre-processed. In this tutorial, we will first summarize our main
recommendations for the handling and preprocessing of point-cloud data,
and then explain how to use compete_pc() and the associated
TreeCompR functions to compute competition indices.
To illustrate how TreeCompR can be included inside a
tidy workflow, we will make use of tidyverse functions throughout the
tutorial:
To highlight where they are coming from, we will explicitly quote the
corresponding package for all functions used in the tutorial in the form
of purrr::map() for all functions besides the
magrittr pipe operator %>%.
Pre-processing the point clouds
Before using the cone and cylinder point cloud approaches, the trees for which competition is to be quantified must be extracted from the plot. The most accurate (if time consuming) way to do this is by manual segmentation, for instance with CloudCompare. Make sure that the point cloud is ready to use (e.g. co-registration of the TLS scans, or any filtering of outliers should be done beforehand).
In CloudCompare, load your point cloud of the whole plot and use the
Segment
function to extract the target tree. Make sure you do not miss any part
or include neighboring trees or their branches, as it is crucial to
determine which parts of the point cloud belong to neighbors and which
points belong to the target tree. Pay particular attention to the ground
and do not include any ground points in your target tree’s point cloud,
otherwise the tree’s base position may be determined imprecisely in
later steps as the tree_pos() function is based on the
lowest points in the point cloud of the target tree.
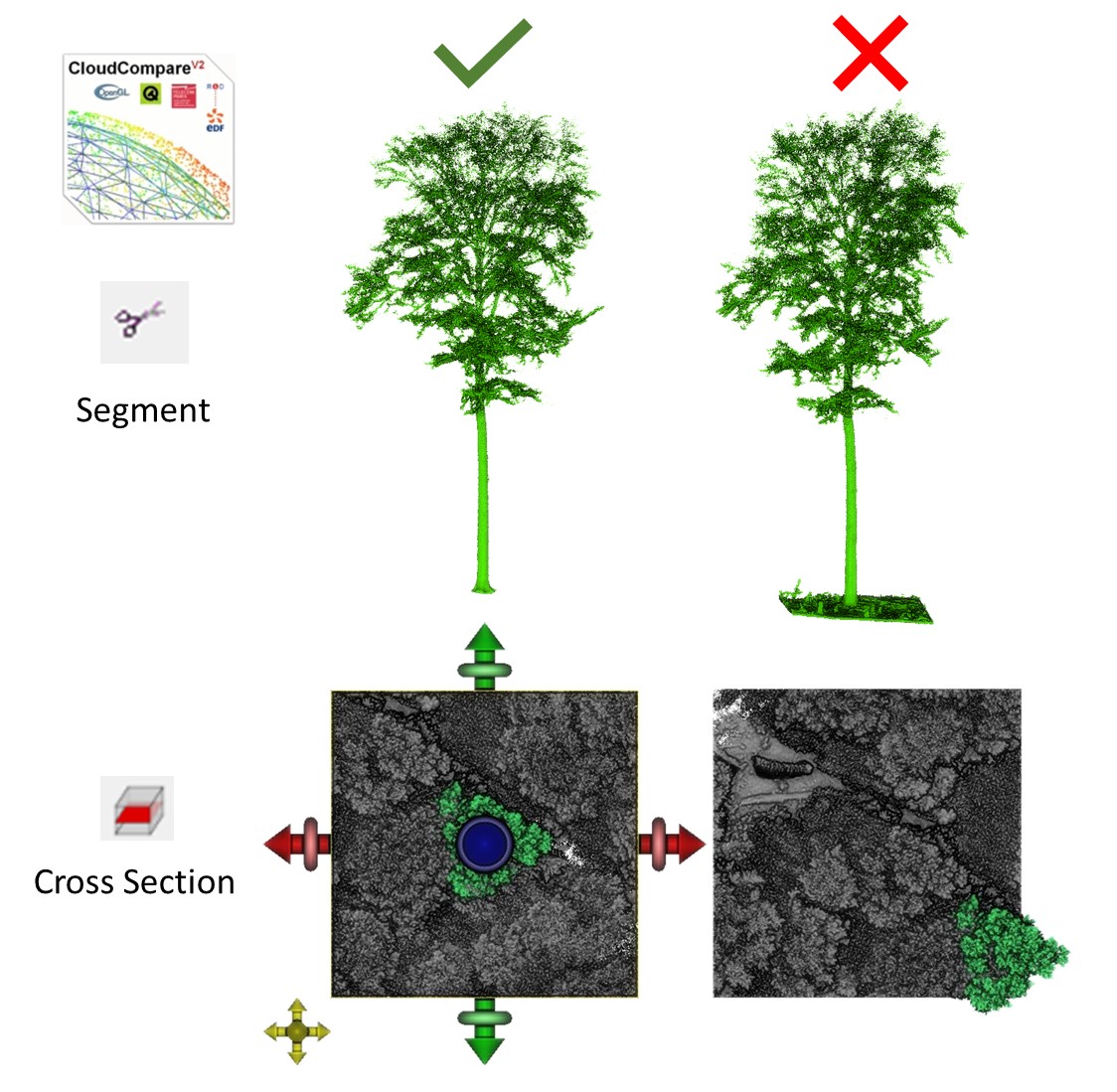
After extracting the target tree, export the point cloud by selecting the file in the DB Tree and click File - Save. Do the same for the whole plot, but first clip the point cloud to your area of interest. The best way to do this is to select top view and use the Cross Section function to clip your point cloud. Make sure that your target tree is not at the edge of the plot. For the example dataset in our paper, we used 30 x 30 m plots.
When exporting/saving the point clouds, you can select .las/.laz, .txt, .csv, … TreeCompR is flexible with file formats, but if possible, use the same format for target trees and their neighborhoods, as there may be slight numeric differences between formats that complicate coordinate matching.
After clicking save, you will be asked to select the output
resolution. If you choose a custom resolution or coordinate accuracy,
make sure you choose the same for the target tree and the neighborhood
cloud! In our examples, we rounded our coordinates to 2 decimal places
(i.e. to 1 cm accuracy), which also is what the
compete_pc() uses by default for coordinate matching, but
other values are possible.
Quantifying competition with TreeCompR
Once you have the two point clouds for target tree and neighborhood,
you can use the read_pc() function of TreeCompR to read and
validate them, or directly pass the paths to the point clouds to
compete_pc().
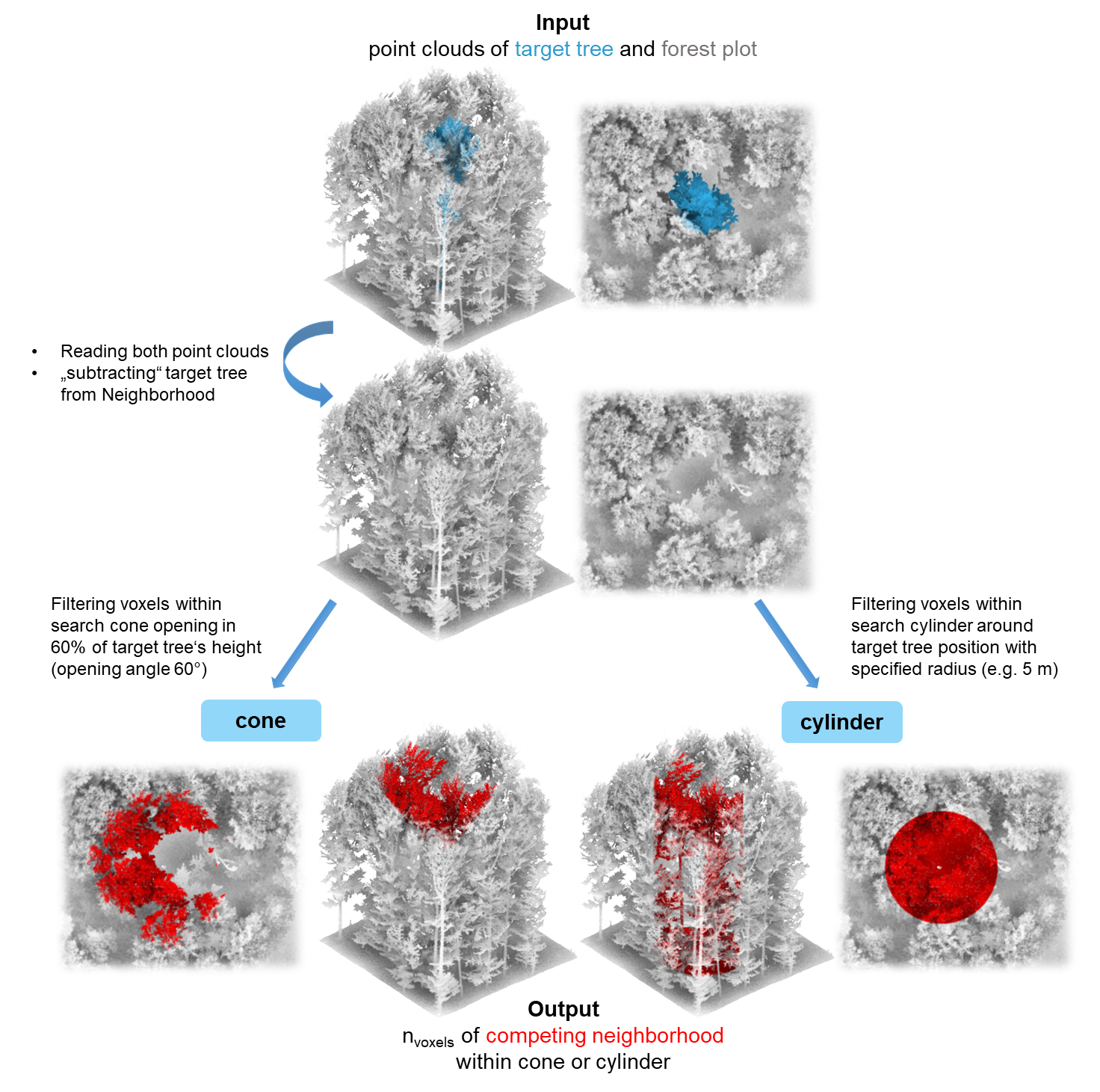
Basic use of compete_pc()
In compete_pc(), you can select the method for computing
competition indices by setting comp_method to
"cylinder", "cone" or "both". To
compute the competition indices for either of these methods, the
position of the center of the cone or cylinder has to be defined. The
center_position argument allows to center the cone or
cylinder either around the central point of the crown projected area
("crown_pos") or the central point of the tree base
("base_pos"). Usually, the position of a tree is calculated
by using the lowest points of a tree and take the mean or median.
However, if a tree is not growing straight and we want to identify
(crown-)competition, we think it is more reasonable to instead focus on
the center of the crown. For that reason, compete_pc()
currently defaults to center_position = "crown_pos", the
median of the x and y position of the voxelized crown projected area
(see the documentation of tree_pos() for details).
If you want to use the stem base position, there are additional
parameters that you can adjust to ensure the correct position
determination, namely h_xy (range in m above the lowermost
point of the tree point cloud the base position should be calculated
from) and z_min (minimum number of points in the lowermost
layer of voxels used to calculate base position of the tree). More about
the methods themselves and the settings and sensitivity can be found in
Seidel et
al. (2015), Metz
et al. (2013), or, for the original KKL index, in Pretzsch
et al. (2002).
In the following example, the compete_pc is used to calculate both cone and cylinder-based competition indices centered around the crown center, with a cone opening in a height of 0.6 times the total target tree height and a cylinder radius of 4 m:
# compute competition indices
CI1 <- compete_pc(
forest_source = "data/neighborhood.txt",
tree_source = "data/tree.txt",
comp_method = "both", # calculate CI in cylinder and cone
center_position = "crown_pos", # center for cone and cylinder
cyl_r = 4, # radius in m
h_cone = 0.6, # position where the cone starts to open relative to the
# height of the target tree (0.6 = 60 % of tree height)
print_progress = "some" # controls how much of the progress is
) # printed
#> ----- Processing competition indices for: tree -----
#> Cone-based CI = 15330 Cylinder-based CI = 55815 With the standard settings (print_progress = "some"),
the function only prints the computed competition indices. When no
output is desired, the argument can be set to "none", when
even more detail are required it can be put to "full".
Having some idea whether the computed values are reasonable is rather
useful when looping over a large number of trees as the computations can
become rather time-consuming for large datasets.
The resulting compete_pc object looks like this:
CI1
#> -------------------------------------------------------------------
#> 'compete_pc' class point-cloud based competition indices for 'tree'
#> -------------------------------------------------------------------
#> target height_target center_position CI_cone h_cone CI_cyl cyl_r
#> <char> <num> <char> <int> <num> <int> <num>
#> 1: tree 22.8 crown center 15330 0.6 55815 4The result is an object of type compete_pc, i.e. a
modified data.frame that you can save or use for further statistical
analysis. It contains the name or ID (filename) of the tree, its height,
the type center_position used, the computed competition index value(s)
(voxel counts) and the settings of the parameters controlling cylinder
and cone size.
Reading different formats with read_pc()
If you want to use the same neighborhood again because you have more
than one target tree within that plot, you do not need to read the point
cloud from the file each time. Instead, simply read it once with
read_pc() and use it again. You can also load the target
tree outside the compete_pc() function.
neighbours <- read_pc("data/neighborhood.txt")
#> No named coordinates. Columns no. 1, 2, 3 in raw data used as x, y, z
#> coordinates, respectively.
tree <- read_pc("data/tree.txt")
#> No named coordinates. Columns no. 1, 2, 3 in raw data used as x, y, z
#> coordinates, respectively.If reading plain-text formats like .txt or .csv in that form, you will be notified about the columns identified as coordinates (any capitalization of x, y and z, or the first 3 numeric columns if that is not possible).
These objects can then be passed to compete_pc() to
calculate competition indices:
# compute indices
CI2 <- compete_pc(
forest_source = neighbours, tree_source = tree,
comp_method = "both", center_position = "crown_pos",
cyl_r = 4, h_cone = 0.6, print_progress = "none"
)If required, non-standard decimal and field separators (as well as
other inputs) can be passed to data.table::fread() to be
able to read more exotic formats:
tree0 <- read_pc("data/tree0.csv", sep = ";", dec = ",")In most cases, the data will be available in a data format dedicated
specifically for the storage of laser scanning data. In addition to all
plain-text formats readable to data.table::fread(),
read_pc() and compete_pc() can both read .las,
.laz and .ply formats:
# Read a tree point cloud in .las format
tree1 <- read_pc(pc_source = "data/tree_point_cloud.las")
# Read a tree point cloud in .ply format
tree2 <- read_pc(pc_source = "data/tree_point_cloud.ply")
# compute competition with source files in .laz format
CI3 <- compete_pc(
forest_source = "data/neighbors1.laz", tree_source = "tree1.laz",
comp_method = "cone",
print_progress = "none"
)Data handling in read_pc() and
compete_pc()
Internally, compete_pc() crops the neighborhood to the
immediate surroundings of the cone or cylinder before matching the
neighbor and tree point clouds, as for large neighborhoods this is the
by far computationally most expensive step. Once a neighborhood is
loaded, filtering (which is done efficiently using
data.table functions package) is comparably fast. To speed
up the coordinate matching, it is done on integer coordinates rounded to
the digit accuracy specified by acc_digits, which defaults
to 2 (i.e., data are by default rounded to 2 digits, i.e. the nearest
cm). Duplicate observations are then discarded already during the
read_pc() step to reduce memory load and speed up
computations. This built-in downsampling ensures consistency in
coordinates between target and neighbor trees (as long as they come from
a coordinate system with the same origin, which is a prerequisite for
the analysis).
Should you have substantially more accurate data, you can consider
increasing that value, though it will slow down computations and likely
will not have a large impact on the outcome as the competition indices
are computed on a coarser spatial scale set via the res
argument in compete_pc() (by default,
res = 0.1, i.e. the analysis itself is performed with
voxels with 0.1 m edge length). The number of digits can be modified as
follows:
# Read a tree point cloud with 4 digits accuracy
acc_tree <- read_pc(pc_source = "data/tree_point_cloud.las", acc_digits = 4)If a forest_pc object is used as an input for
compete_pc, the function maintains the accuracy of the
imported object (which is stored as attr(x, "acc_digits")),
and it is tested if the acc_digits for target tree and
neighborhood is consistent.
Since loading a large point cloud into memory has a non-negligible computational overhead and can take a lot of time (especially on systems with older hard drives), loading a neighborhood just once can speed up the analysis considerably when batch processing a large number of trees from the same data source even if the neighborhood is large. Should the objects become too large to handle within memory, we recommend to split them up outside R during the data preparation step to ensure each subset contains one part of the forest with one or several target trees.
Example for batch-processing workflows
Working with a lookup table
If your study consists of many target trees that each are surrounded by their own neighborhood plot, it may be useful to create a lookup table to more efficiently run the function for all your trees. Here is an example of what such a lookup table might look like:
# Define the base paths
neighborhood_path <- "data/neighborhood/"
trees_path <- "data/trees/"
# List .las files in the neighborhood and tree folders
neighborhood_files <- list.files(neighborhood_path, full.names = TRUE,
pattern = "\\.las$")
tree_files <- list.files(trees_path, full.names = TRUE, pattern = "\\.las$")
# Create TreeIDs by stripping the file extension from the tree files
TreeIDs <- gsub("\\.las", "", list.files(trees_path, full.names = FALSE,
pattern = "\\.las$"))
# Create the lookup table
lookup_table <- tibble::tibble(
tree_name = TreeIDs,
forest_source = neighborhood_files,
tree_source = tree_files,
stringsAsFactors = FALSE
)The table should now look like this:
lookup_table
#> tree_name forest_source tree_source
#> <chr> <chr> <chr>
#> 1 YW01 data/neighborhood/YW01_neigh.las data/trees/YW01.las
#> 2 DK01 data/neighborhood/DK01_neigh.las data/trees/DK01.las
#> 3 GN01 data/neighborhood/GN01_neigh.las data/trees/GN01.las
#> 4 AR01 data/neighborhood/AR01_neigh.las data/trees/AR01.las
#> 5 BS01 data/neighborhood/BS01_neigh.las data/trees/BS01.las
#> 6 YW02 data/neighborhood/YW02_neigh.las data/trees/YW02.las
#> 7 DK02 data/neighborhood/DK02_neigh.las data/trees/DK02.las
#> 8 GN02 data/neighborhood/GN02_neigh.las data/trees/GN02.las
#> 9 AR02 data/neighborhood/AR02_neigh.las data/trees/AR02.las
#> 10 BS02 data/neighborhood/BS02_neigh.las data/trees/BS02.las
#> # ℹ 15 more rowsIn this case, the lookup table contains one path for each segmented target tree point cloud, and another path for the corresponding 30 x 30 m² neighborhood.
We can now use dplyr::mutate() to append the other
function arguments for compete_pc(), and
purr::pmap() to map over the paths and settings line by
line and use compete_pc() with the settings specified in
each of them. As this can take a considerable amount of time if
processing hundreds of trees, the different options for
print_progress can be useful to be able to keep track of
the analysis and interrupt it in time if something goes wrong.
#define additional parameter settings for compete_pc within the lookup table
lookup_cone50_cyl5 <- lookup_table %>%
dplyr::mutate(comp_method = "both",
center_position = "crown_pos",
cyl_r = 5, h_cone = 0.5,
z_min = 100, h_xy = 0.3,
print_progress = "some")
# use pmap to loop over the table
results <- lookup_cone50_cyl5 %>%
purrr::pmap(compete_pc)
#> ----- Processing competition indices for: YW01 -----
#> Cone-based CI = 15046 Cylinder-based CI = 101332
#>
#> ----- Processing competition indices for: DK01 -----
#> ... <full console print not shown> ...By printing the process tree by tree, it is possible to estimate the total time it takes to compute the indices for large datasets and identify in time when the function gets stuck.
It is possible to bind the results for several trees into a single dataset using bind_rows:
results_table <- bind_rows(results)
results
#> -----------------------------------------------------------------------------
#> 'compete_pc' class point-cloud based competition indices for 25 target trees
#> -----------------------------------------------------------------------------
#> target height_target center_position CI_cone h_cone CI_cyl cyl_r
#> <char> <num> <char> <int> <num> <int> <num>
#> 1: YW01 21.8 crown center 15046 0.5 101332 5
#> 2: DK01 18.5 crown center 16654 0.5 130674 5
#> 3: GN05 24.2 crown center 13142 0.5 125293 5
#> ---
#> 23: GN05 23.9 crown center 13252 0.5 139834 5
#> 24: AR05 20.8 crown center 17651 0.5 150293 5
#> 25: BS05 22.1 crown center 11306 0.5 114520 5Multiple trees in one plot
If you have more than one tree in the same neighborhood, it makes sense to load the neighborhood first and then loop over the trees to avoid loading the neighborhood several time:
# Read neighborhood
neighbor <- read_pc("data/neighborhood1.las")
# read trees
trees <- list.files("data/trees1", pattern = "\\.las$")
# use purrr::map() to compute all trees
results1 <- map(
trees,
~compete_pc(
forest_source = neighbor,
tree_source = file.path("data/trees1", .x),
cyl_r = 4,
tree_name = gsub("\\.las", "", .x), # take id directly from name
comp_method = "cylinder")
) %>%
bind_rows()If there is overlap in the studied areas, this may be faster than loading local subsets separately for each tree even for relatively large neighborhoods because loading the data tends to be slower than processing them once they are in memory.
Dealing with high point densities
Point clouds obtained from static ground-based scanners (TLS) can have very high point densities, sometimes resulting in file sizes that make it impossible to load the raw point cloud into memory in R. Whenever possible, the point cloud should be clipped beforehand, ideally as a pre-processing step in dedicated software for these methods such as CloudCompare.
If this is not sufficient, the point cloud can be reduced in size,
e.g. by downsampling or by voxelizing the point cloud. Depending on the
method, however, this may affect the results. In
read_inv(), we implemented a simple and fast reduction of
data size by rounding all coordinates to a coordinate accuracy specified
by acc_digits using Rfast::Round(). While the
same can also be achieved in R outside TreeCompR, different
solutions such as lidR::voxelize_points() or
VoxR::vox() may result in different results depending on
the chosen origin of the voxel grid. For these reasons, we prefer to do
this step in our package in a consistent fashion that permits internal
checks of coordinate accuracy and allows to match the target tree and
neighborhood pointclouds based on integer coordinates to speed up
calculations and avoid falling into The Floating Point Trap (See chapter
1 of the R Inferno by Burns, 2001).
This combination of pre-processing and built-in data aggregation
should be sufficient for a majority of datasets. If the raw data are too
large to be read into memory even after clipping to the surroundings of
the target tree, it makes sense to further reduce their size by
voxelizing/rounding before loading, e.g. in CloudCompare. Here,
ideally round to 1-2 decimal places more than desired
acc_digits, because the storage format may result in slight
numeric differences (especially for .las/.laz) that can affect
coordinate matching.
Downsampling along a voxel-based grid such as in
TreeLS::smp_voxelize() is strongly discouraged for this
purpose, as the subsampled points will not match between neighborhood
and target tree point cloud, and may not even have the same coordinates
after converting them into voxels depending on the origin of the voxel
grid.